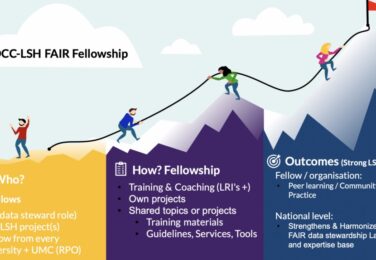
Interoperability Challenges and Recommendations Across SSH, LSH, and NES
Summary of feedback received at the Open Science Festival 2025, Groningen
Authors: Walter Baccinelli, Alex Brandsen, Vedran Kasalica, Rory Macneil & Errol Neo
The three TDCCs ran a joint workshop at the Open Science Festival on interoperability, to help the newly hired interoperability experts to get an idea of the challenges and recommendations in our respective domains. After an introduction by Rory Macneil, we split into breakout groups for each domain. Below is an overall summary, followed by more detailed information about each domain breakout.
Image by Scriberia, via the Turing Way
Overall summary
Across SSH, LSH, and NES, researchers face similar interoperability challenges rooted in cultural, technical, and educational gaps. Interoperability is often poorly understood, undervalued, and introduced too late in the research process. Technical barriers include fragmented infrastructures, inconsistent or missing metadata and ontologies, siloed or proprietary data formats, and limited visibility or sustainability of open tools. Researchers frequently encounter vendor lock-in, incompatible systems, and poorly documented software, all of which hinder data reuse and reproducibility.
Culturally, incentives are weak: mandates alone do not motivate researchers, and good practices are not consistently rewarded or shared. Many solutions remain local due to limited peer learning and unclear guidance.
Participants across domains agreed that interoperability must be built into the full research lifecycle and supported by better training, clearer standards, open and discoverable tools, and stronger institutional support. They envision a future with seamless workflows, shared frameworks, practical use-case-driven methods, and roles or systems dedicated to maintaining interoperability.
SSH summary
The discussion identified multiple, interconnected barriers to achieving FAIR interoperability in SSH research. Participants stress that many researchers lack a shared understanding of interoperability itself, and that awareness and training are missing from BA/MA education and the wider research culture. Infrastructure is fragmented or insufficient, with gaps in open-source tools, semantic resources, and support for qualitative data. Ontologies differ across subfields and are often not explicit or findable, which hampers cross-disciplinary reuse.
Practical constraints also arise from vendor lock-in, software version dependencies, and divergent data management systems across organisations, which complicate code reproducibility and cross-platform data exchange. Sustainability issues affect persistent identifiers: long-term maintenance and versioning remain unresolved concerns.
Participants emphasised that interoperability enters research too late. They argue it should be embedded across the entire lifecycle, supported by clearer guidelines, best-practice examples, and peer learning opportunities. Structural incentives are also needed, for example through graduate school requirements or recognition and rewards in career or funding assessment.
LSH summary
During the workshop on interoperability, participants explored concrete cases of interoperability challenges, reflected on possible shared solutions, and discussed how to ensure that experiences and good practices are more widely shared across the community.
1. Where interoperability issues arise
Participants reported a wide range of interoperability issues that affect their daily research and data management work. A recurring theme was the low quality or lack of (meta)data annotations, which makes it difficult to combine datasets from multiple sources. Even when data are available, the lack of consistent ontologies and the high technical barrier to implementing them hinder harmonization efforts.
Another major source of frustration came from proprietary software. Many tools rely on closed data standards and undocumented data models, which severely limit the ability to reuse, share, or integrate data across systems. The absence of open APIs and open-source alternatives further compounds the problem, creating technical and legal lock-ins.
Participants also pointed to infrastructural barriers, especially regarding the sharing of large datasets. The lack of common repositories or standards for storing and exchanging microscopy data makes collaborative work cumbersome.
Finally, additional factors such as unclear data ownership, operating system incompatibilities, and poor documentation exacerbate the interoperability gap, often leaving researchers to reinvent solutions locally.
2. Possible approaches and common solutions
Despite these challenges, participants identified several promising avenues for improvement. One strategy is to learn from other domains that have successfully tackled interoperability. Established frameworks like BBMRI Biobank standards or the ISA Framework, which includes metadata for sample collection and processing, could offer valuable models for adaptation to other disciplines.
Another key proposal was to develop and promote open software alternatives to proprietary tools. Participants emphasized the importance of advocating for legal frameworks that would compel proprietary software providers to make their models and formats open, or at least interoperable. Joint licensing or collective negotiation among institutions could also increase leverage when dealing with vendors, ensuring more consistent and compatible choices across the community.
In addition, participants highlighted the need to increase the findability and visibility of open solutions. Many open tools already exist but are not easily discoverable or evaluated for quality. A dedicated repository that maps existing research software, its interoperability features, and its maturity could serve as a valuable community resource.
3. Sharing problems and solutions
To sustain progress, participants stressed the importance of sharing both challenges and solutions in accessible and engaging ways. One idea was to create “spotlights” that showcase existing interoperability solutions and the real-world issues they helped to overcome. Another creative suggestion was to develop a “horror stories” series, where researchers can share their experiences with broken workflows, inaccessible data, or incompatible software—helping others learn from these pitfalls.
Finally, embedding accessible citation metadata directly into platforms like ORCID could facilitate recognition and reuse of interoperability efforts, making it easier to trace and credit contributions to open and interoperable science.
NES summary
During the session, participants were asked what interoperability challenges they faced, and what they wished to see in interoperability development in the NES domain in the next 5 years.
Part 1: Challenges - What is preventing us from making outputs more interoperable?
Participants identified three main categories of obstacles: Cultural/Motivational, Technical/Structural, and Educational.
- Cultural & Motivational Barriers
- Lack of Incentives: There is a struggle to motivate researchers to improve data interoperability. Data stewards shared a sentiment that researchers question why they should change their workflows. Many do not care about interoperability or view it as an administrative burden.
- Mandates vs. Motivation: While institutes provide mandates, they are not always sufficient to drive genuine engagement without demonstrating value through specific use cases.
- Technical & Structural Issues
- Siloed Data: Data is often not interoperable, even between different departments within the same institute.
- Proprietary Constraints: Data generated by lab machines and specific software often lock users into formats that are not interoperable by design.
- Tool Visibility: There is currently a lack of a clear map of tools used throughout the research workflow, making take-up and integration difficult.
- System Usability: Current systems and repositories are not self-explanatory; moreover, better indexing on publishing platforms is required.
- Knowledge Gaps & Support
- Unfamiliarity with Ontology: Students and early-career researchers often find ontology concepts unclear.
- Lack of Tooling: There is a distinct need for templates and tools to assist in making data interoperable.
- Repository Checks: There is a desire for repositories to play a more active role in checking for interoperability upon data input.
Part 2: Wishes - What do we wish to see in the NES domain by 2030?
The discussion outlined a future where interoperability is integrated into the research lifecycle rather than treated as an afterthought.
- Standardization & Automation
- Knowledge of Formats: A future where all researchers have learned which preferred data formats to use, eliminating the need for manual conversion later.
- Workflow Interoperability: Seamless connection between different research services and tools, creating a unified workflow.
- Structural Support & Personnel
- Interoperability Officers: A vision where every research project involves an interoperability officer to offer advice.
- Strategic Prioritization: A clear understanding of the different "levels" of interoperability—identifying which levels require flexibility and which are critical to tackle immediately.
- Methodology
Clarity: Methods to improve interoperability should be standardized and easy to pick up once commonalities are identified.
Use-Case Driven: Moving away from abstract concepts to concrete use cases that highlight similarities across disciplines.